Testing for differences between two mean spectra and results:
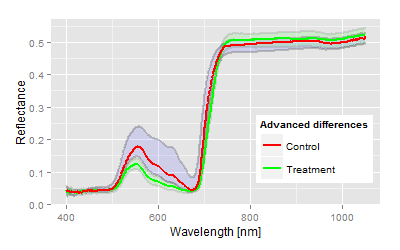
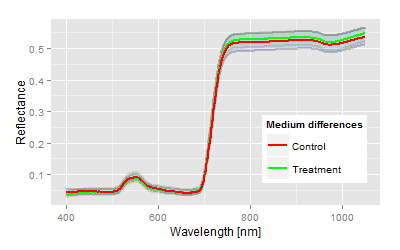
We have chosen one top and bottom end example to demonstrate the analysis of a two means comparison problem, somehow equivalent to a t-test (if you ignore the assumptions of a t-test). The graphs show the mean reflectance signatures of both control and treatment with the 95% confidence bands in the domain up to 1050 nm.| Visualisation: Spectral example for highly significant differences |
Visualisation: Spectral example for no differences (n.s.) |
 |
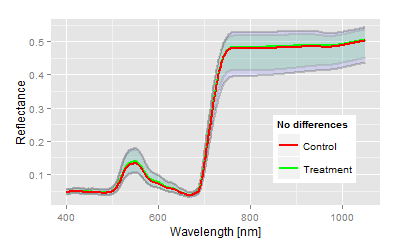 |
After model fitting and discriminant analysis the result contains several statistical parameter (showing simultaneously in the same direction in most of the cases), which allow the interpretation of the trial. The interpretation must not be overstressed, the question in this case:
Is there a treatment effect compared to the control?
Nothing more and nothing less.
| The table summarises the two examples depicted in the graphs above in terms of their gained statistical parameters, how these parameter changes with the situation, and what kind of interpretation exist. | ||||
| Statistical parameter | Different | Explanation | No difference | Explanation |
| c2 | P=0.000 | The significance of the discriminant functions gives a first hint of treatment effects. As smaller the probability value p, as larger the differences | p=0.924 | The usual boundaries are used: p>0.05 or better p>0.01, non significant (n.s.) |
| Canonical correlation | r=0.93 | As in the classical correlation we can use the common separation:
|
r=0.193 | no correlation, not significant |
| Canonical distance | 4.9 | The most interesting parameter: Based on an arbitrary, dimensionless scale we can quantify the comparison, the value describes the intensity of the treatment effect. | 0.4 | Distance is less than 1, obvious no treatment effect. |
| Classification | 95% | Very high percentage of correct classification, the data distribution indicates a large difference and a clear distinction. | 50% | The bottom end of any classification; the result indicates randomness; no distinction possible; in fact no treatment was induced in this data set. |
ANOVA of the mean comparison |
||||||
| Difference | No difference | Explanation | ||||
| Parameter | Model label | F-value | p-value | F-value | p-value | 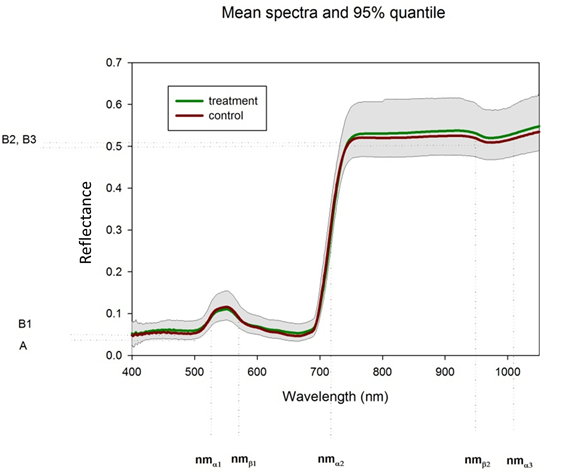 Slight cut backs must be made in the interpretation of the ANOVA table, as the internal correlation of the
model parameter is not taken into account in the ANOVA, therefore it should be used for orientation only. The
standard boundary of 5% for the statistical decision is not valid here.
We assume p-values <0.01 (or < 1%) are significant, certainty exists with p-values <0.000. The more the
spectra differ, the more model
parameters are significant different. In case of significances, the size of the F-value depicts how
much of the distinction is affected by this parameter and in which domain of the spectra treatment effects
are largest. The model exponents (not shown in the graph) are describing the slopes of the amplitudes, but
should not get too much weight alone in the interpretation of the results.
Slight cut backs must be made in the interpretation of the ANOVA table, as the internal correlation of the
model parameter is not taken into account in the ANOVA, therefore it should be used for orientation only. The
standard boundary of 5% for the statistical decision is not valid here.
We assume p-values <0.01 (or < 1%) are significant, certainty exists with p-values <0.000. The more the
spectra differ, the more model
parameters are significant different. In case of significances, the size of the F-value depicts how
much of the distinction is affected by this parameter and in which domain of the spectra treatment effects
are largest. The model exponents (not shown in the graph) are describing the slopes of the amplitudes, but
should not get too much weight alone in the interpretation of the results. |
| A | A | 0.821 | 0.371 | 0.097 | 0.756 | |
| B1 | B1 | 25.717 | 0.000 | 0.000 | 0.994 | |
| nma1 | AC1 | 0.408 | 0.527 | 0.250 | 0.617 | |
| nmb1 | BC1 | 39.282 | 0.000 | 0.41 | 0.840 | |
| a1 | AL1 | 4.134 | 0.049 | 0.451 | 0.503 | |
| b1 | BE1 | 3.060 | 0.089 | 0.474 | 0.492 | |
| B2 | B2 | 16.369 | 0.000 | 0.008 | 0.930 | |
| nma2 | AC2 | 62.845 | 0.000 | 0.011 | 0.920 | |
| nmb2 | BC2 | 37.501 | 0.000 | 0.003 | 0.054 | |
| a2 | AL2 | 37.778 | 0.000 | 0.041 | 0.839 | |
| b2 | BE2 | 4.856 | 0.034 | 0.198 | 0.657 | |
| B2 | B3 | 16.869 | 0.000 | 0.003 | 0.955 | |
| nma3 | AC3 | 50.742 | 0.000 | 0.343 | 0.558 | |
| a3 | AL3 | 15.492 | 0.000 | 0.375 | 0.541 | |
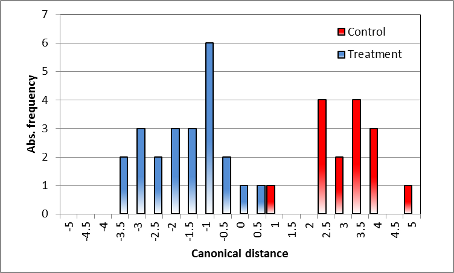
Summary classification | |
| Distribution of classification (discriminant scores), different | Distribution of classification (discriminant scores), no difference |
 |
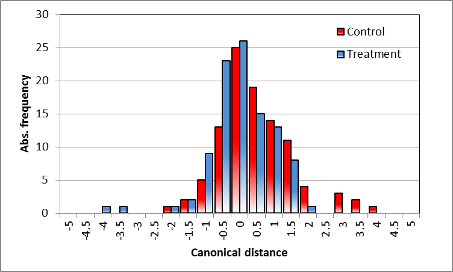 |
| Demonstration of the variance found in the spectral measurements, also affected by the individuality of the specific single plant. No overlapping exits in the distribution, but an obvious discrimination of the treatment. | Obvious overlapping of the distance classes, no distinction of the spectra, variance larger than the mean canonical distance. |
A typical outcome of an experiment
On average data will be most likely in between the two extremes as the following example shows; the following spectra were recorded with given variance
| Spectra | Distribution of discriminant scores |
 |
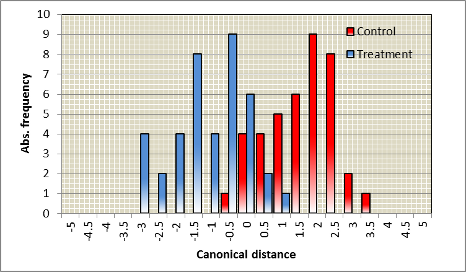 |
| Statistical parameter | Value | Explanation |
| c2 | P=0.000 | Discriminant function is significant indicating a significant difference between treatment and control. |
| Canonical correlation | 0.80 | Correlation is high on a scale from 0 to +1, also for this parameter, the difference is obvious. |
| Canonical distance | 2.74 | The averaged distance is relatively high, overlapping classification scores are due to the natural variance by the individual plant. |
| Classification | 90% | Extremely high for spectra apparently not differing from each other. |
| Model label | F-value | p-value | ANOVA table and explanation |
| A | 0.331 | 0.567 | By chance, the given example demonstrate the limits of the ANOVA. Numerous parameter are below a p-Value of 5%, but by the criteria to use here only parameter Bcrit2 and Acrit3 account for the differences caused by the treatment. Both parameter describes the amplitude in the higher wavelength domains. The treatment effect leads to changes in the structural tissue of the plant. |
| B1 | 0.072 | 0.790 | |
| AC1 | 2.858 | 0.095 | |
| BC1 | 4.041 | 0.048 | |
| AL1 | 4.962 | 0.029 | |
| BE1 | 6.362 | 0.014 | |
| B2 | 4.389 | 0.039 | |
| AC2 | 0.004 | 0.951 | |
| BC2 | 7.248 | 0.009 | |
| AL2 | 8.362 | 0.005 | |
| BE2 | 0.099 | 0.754 | |
| B3 | 4.478 | 0.038 | |
| AC3 | 30.832 | 0.000 | |
| AL3 | 1.173 | 0.282 |
 Evaluation reinvented
Evaluation reinvented
